کیت استخراج Total RNA Isolation kit | RNA
15,800,000 ریال
توضیحات محصول
کیت استخراج Total RNA Isolation kit | RNA
MEGARENA
High Pure Total RNA extraction kit
Cat. No:
FPKT029.0025
FPKT029.0050
FPKT029.0100
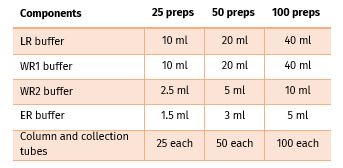
Contents:
Kit storage:
 This kit should be stored at room temperature.
This kit should be stored at room temperature.
 If properly stored, all kit components are stable until the expiration date printed on the label.
If properly stored, all kit components are stable until the expiration date printed on the label.
Additional Equipment and Reagent required
- Make sure everything is RNase-free when handling RNA.\• Absolute ethanol
- ß-Mercaptoethanol
- Standard tabletop microcentrifuge capable of 13,000 x g centrifugal force
- Microcentrifuge tubes, 1.5 ml, sterile
For Isolation of Total RNA from Cultured Cells:
- PBS
For the isolation of total RNA from human blood:
- Red Blood Cell Lysis Buffer (RLT)
- PBS
For the isolation of total RNA from Animal Tissue:
- liquid nitrogen & mortar
- a rotor-stator homogenizer or a 20-G needle syringe
For the isolation of total RNA from yeast:
- Lyticase or zymolase
- PBS
For the isolation of total RNA from bacteria:
- Lysozyme (Lysozyme reaction solution: (10mg/ ml lysozyme; 20mM Tris-HCl, pH 8.0; 2mM EDTA;1.2% Triton))
For the isolation of Total RNA from Paraffin-embedded tissue
- xylene
Application
The High Pure RNA Isolation Kit is designed for the purification of total RNA from cultured cells. Other sample materials like Animal Tissue, blood, yeast, and bacteria require an additional specific pre-lysis treatment, which is described in the Additional Equipment and Reagent required. RNA is suited for other techniques like northern blotting, RNase protection, and primer extension. The procedure is optimized to achieve reliable results within Approximately 1 hour (24 samples simultaneously).
Handling Requirements and Safety Information
 All solutions are clear and should not be used when precipitates have formed. Warm the solutions at +15 to +25°C or in a 37°C water bath until the precipitates have dissolved.
All solutions are clear and should not be used when precipitates have formed. Warm the solutions at +15 to +25°C or in a 37°C water bath until the precipitates have dissolved.
 LR Buffer and WR1 Buffer contain guanidinium hydrochloride which is an irritant.
LR Buffer and WR1 Buffer contain guanidinium hydrochloride which is an irritant.
 ß-Mercaptoethanol (ß-Me) is hazardous to human health. perform the procedures involving ß-Me in a chemical fume hood.
ß-Mercaptoethanol (ß-Me) is hazardous to human health. perform the procedures involving ß-Me in a chemical fume hood.
 Do not allow LR Buffer and WR1 Buffer to touch your skin, eyes, or mucous membranes. If contact does occur, wash the affected area immediately with large amounts of water.
Do not allow LR Buffer and WR1 Buffer to touch your skin, eyes, or mucous membranes. If contact does occur, wash the affected area immediately with large amounts of water.
 If you spill the reagent, dilute the spill with water before wiping it up.
If you spill the reagent, dilute the spill with water before wiping it up.
 Do not use any modified ethanol.
Do not use any modified ethanol.
 Do not pool reagents from different lot numbers.
Do not pool reagents from different lot numbers.
 Immediately after usage, close all bottles to avoid leakage, varying buffer concentrations, or buffer conditions.
Immediately after usage, close all bottles to avoid leakage, varying buffer concentrations, or buffer conditions.
 After first opening store all bottles in an upright position.
After first opening store all bottles in an upright position.
 Do not allow the LR Buffer and WR1 Buffer to mix with sodium hypochlorite found in commercial bleach solutions. This mixture can produce highly toxic gas.
Do not allow the LR Buffer and WR1 Buffer to mix with sodium hypochlorite found in commercial bleach solutions. This mixture can produce highly toxic gas.
 Wear protective disposable gloves, laboratory coats, and eye protection, when handling samples and kit reagents.
Wear protective disposable gloves, laboratory coats, and eye protection, when handling samples and kit reagents.
 Do not contaminate the reagents with bacteria, viruses, or nucleases. Use disposable pipets and nuclease-free pipet tips to remove aliquots from reagent bottles.
Do not contaminate the reagents with bacteria, viruses, or nucleases. Use disposable pipets and nuclease-free pipet tips to remove aliquots from reagent bottles.
preparation procedure:
Working Solution Preparation
FPKT033.0025-25 prep
 Add 2.5 ml absolute ethanol to WR1 Bottle.
Add 2.5 ml absolute ethanol to WR1 Bottle.
 Add 10 ml of absolute ethanol to WR2 Bottle.
Add 10 ml of absolute ethanol to WR2 Bottle.
FPKT033.0050-50 prep
 Add 5 ml absolute ethanol to WR1 Bottle
Add 5 ml absolute ethanol to WR1 Bottle
 Add 20 ml absolute ethanol to WR2 Bottle
Add 20 ml absolute ethanol to WR2 Bottle
FPKT033.0100-100 prep
 Add 10 ml of absolute ethanol to WR1 Bottle.
Add 10 ml of absolute ethanol to WR1 Bottle.
 Add 40 ml of absolute ethanol to WR2 Bottle.
Add 40 ml of absolute ethanol to WR2 Bottle.
 Please note that the Ethanol concentration of a Washing Buffer may decrease during long-term storage resulting in a drop-down of the final RNA yield.
Please note that the Ethanol concentration of a Washing Buffer may decrease during long-term storage resulting in a drop-down of the final RNA yield.
 before starting Incubate the ER buffer at 55 to 65 ° C until the end of the protocol to obtain the maximum yields.
before starting Incubate the ER buffer at 55 to 65 ° C until the end of the protocol to obtain the maximum yields.
Protocols
Isolation of Total RNA from Cultured Cells (suited for 1~ 5× 106 cells)
- Collect 1 ~ 5 ×106 cells by centrifuge at 300 x g for 5 min at 4 °C. Remove all the supernatant.
- Resuspend cells in 200 μl PBS.
 Do not overload, too much sample will make cell lysis incomplete and lead to lower RNA yield and purity.
Do not overload, too much sample will make cell lysis incomplete and lead to lower RNA yield and purity.
- Add 400 μl LR Buffer and 3.5 μl of ß-Mercaptoethanol and Vortex vigorously for 1 min to resuspend the cells completely.
- Add 600 μl Absolute ethanol to the homogenized lysate, and mix well by pipetting.
 If the clump is still visible after the vortex, pipet the sample mixture up and down to break down the clump.
If the clump is still visible after the vortex, pipet the sample mixture up and down to break down the clump.
- To transfer the sample to a High Pure Filter Tube:
– Insert one High Filter Tube in one Collection Tube.
– Pipet the entire sample into the upper reservoir of the Filter Tube (max. 800 μl). Insert the entire High Pure Filter Tube assembly into a standard tabletop centrifuge. Centrifuge the tube assembly for 1 min at ≥8000 xg (≥10,000 rpm). - Optional Step: DNase I digestion to eliminate genomic DNA contamination follow the step from 6a. Otherwise, proceed to step 7 directly.
6a. Discard the flow through. Add 250 μl of WR1 to the Filter Tube, and centrifuge for 30 sec at ≥8000 xg (≥10,000 rpm).
6b. Discard the flow through. For each sample, pipette 5 µl of 10x DNase incubation buffer into a new sterile microtube and add 5 µl of DNase I.
with DEPC-treated water reach to a volume of 50 µl, then pipette the solution into the upper reservoir of the filter tube. Incubate for 15 min at +15 to +25°C.
6c. Discard the flow through. Add 250 μl of WR1 to the Filter Tube, and centrifuge for 30 sec at ≥8000 xg (≥10,000 rpm).
- Discard the flow through. Add 500 μl WR1 to the upper of the Filter Tube and centrifuge for 30 sec at ≥8000 xg (≥10,000 rpm).
- Discard the flow through. Add 500 μl WR2 to the upper of the Filter Tube. centrifuge 30 sec at ≥8000 xg (≥10,000 rpm). and discard the flow through.
- Leave the tube assembly in the centrifuge and spin it for 3 min at maximum speed (approximately 14,000 rpm) to remove any residual Wash Buffer.
- Discard the Collection Tube and insert the Filter Tube into a clean, sterile RNase-free and DNase-free 1.5 ml microcentrifuge tube.
- Add 50 μl ER Buffer to the upper of the Filter Tube. Centrifuge the tube assembly for 1 min at ≥8000 xg (≥10,000 rpm).
 to obtain the maximum yields you can add the RNA solution to the top of the filter tube and repeat Centrifuge the tube assembly at ≥8000 xg (≥10,000 rpm).
to obtain the maximum yields you can add the RNA solution to the top of the filter tube and repeat Centrifuge the tube assembly at ≥8000 xg (≥10,000 rpm).
 The microcentrifuge tube now contains the eluted Cell RNA. Either use the eluted RNA directly in RT.PCR or store the eluted RNA at −20°C for later analysis.
The microcentrifuge tube now contains the eluted Cell RNA. Either use the eluted RNA directly in RT.PCR or store the eluted RNA at −20°C for later analysis.
Isolation of Total RNA from Human Blood (suited for 200 – 500 μl whole blood)
- Add 1 ml Red Blood Cell Lysis Buffer (RLT: Not Provided) to a sterile 2 ml reaction tube.
- Add 500 μl human whole blood and mix by inversion.Do not vortex.
- Place the tube on a rocking platform or gyratory shaker for 10 min at +15 to +25° C. Alternatively, manually invert the sample periodically for 10 min.
- Centrifuge for 5 min at 500 × g in a standard tabletop centrifuge. With a pipette, carefully remove and properly dispose of the clear, red supernatant.Do not vortex.
- Add 1 ml Red Blood Cell Lysis Buffer (RLT buffer) to the white pellet and mix by “flicking” the tube until the pellet is resuspended.
- Centrifuge for 3 min at 500 × g. Carefully remove and properly dispose of the supernatant, particularly the red ring of blood cell debris that forms around the outer surface of the white pellet.
- Resuspend the white pellet in 200 μl PBS and follow the protocol Isolation of Total RNA from Cultured Cells from step 3.
Isolation of Total RNA from Animal Tissues.
- Weight up to 30 mg of tissue sample. Grind the sample in liquid nitrogen to a fine powder with a mortar and transfer the powder to a new microcentrifuge tube (not provided).
Avoid thawing the sample during weighing and grinding. - Add 350 μl of LR Buffer and 3.5 μl of ß Mercaptoethanol. Homogenize the sample by using a rotor-stator homogenizer or by passing the sample lysate through a 20-G needle syringe 10 times. Incubate at room temperature for 5 min.
- follow the protocol Isolation of Total RNA from Cultured Cells from Step 4.
Isolation of Total RNA from Yeast. (Suited for 1 × 108 cells)
It is recommended to harvest cells during the mid-log or late-log phase of growth (OD600 ≤ 2.0). The cell number can be counted in a hemocytometer chamber or determined by measuring the optical density at 600 nm in a spectral photometer. Use a dilution that gives an A600 of 0.1 – 0.15/ml (0.1 A600 corresponds to approx. 2 ×106cells.)
- Collect the sample by centrifugation at 2,000 × g for 5 min in a standard tabletop centrifuge.
- Add 10 μl Lyticase (0.5 mg/ml), incubate for 15 min at 30°C.
- Follow the protocol Isolation of Total RNA from Cultured Cells from Step 3.
Isolation of Total RNA from Bacteria (gram-positive and gram-negative) (Suited for 1 x 109 cells)
- Collect the sample by centrifugation at 2,000 × g for 5 min in a standard tabletop centrifuge. Resuspend the pellet in 200 μl 10 mM Tris, pH 8.0.
- Add 4 μl Lysozyme (50 mg/ml), incubate for 10 min at 37°C.
- Add 400 μl LR Buffer and mix well
- Combine the High Pure Filter Tube and the Collection Tube and pipette the sample in the upper reservoir.
Centrifuge for 15 s at 8,000 × g in a standard tabletop centrifuge, discard the flow through and again combine the Filter Tube and the used Collection Tube.
- Pipette 40 μl DNase Incubation Buffer into a sterile reaction tube, add 10 μl DNase I, mix and pipette the solution in the upper reservoir of the Filter Tube. Incubate for 60 min at +15 to +25°C.
- Follow the protocol Isolation of Total RNA from Cultured Cells from Step 7.
Isolation of Total RNA from Paraffin-embedded tissue
- Transfer up to 15 mg paraffin-embedded tissue sample to a microcentrifuge tube (not provided). Remove the extra paraffin to minimize the size of the sample slice.
- Add 0.5 ml xylene, mix well, and incubate at room temperature for 10 min.
- Centrifuge at full speed for 3 min. Remove the supernatant by pipetting.
- Add 0.25 ml xylene, mix well, and incubate at room temperature for 3 min.
- Centrifuge at full speed for 3 min. Remove the supernatant by pipetting.
- Repeat step 4 and step 5
- Add 0.3 ml Absolute ethanol to the deparaffined tissue, and mix gently by vortexing. Incubate at room temperature for 3 min.
- Centrifuge at full speed for 3 min. Remove the supernatant by pipetting.
- Repeat step 7 and step 8.
- Follow the Animal tissue Protocol starting from step 1 for sample disruption then follow the protocol Isolation of Total RNA from Cultured Cells from step 4.
RNA Clean-Up Protocol
- Transfer 100 μl of RNA sample to a microcentrifuge tube.
If the RNA sample is less than 100 μl, add RNase-free water to make the sample volume to 100 μl. - Add 300 μl of LR Buffer and 300 μl Absolute ethanol. mix well by vortexing. transfer the sample to a High Pure Filter Tube.
- Centrifuge at full speed for 1 min and discard the flow-through.
- Follow the protocol Isolation of Total RNA from Cultured Cells from Step 7.
بررسی تخصصی
| تعداد Preparation را انتخاب کنید | 100, 50, 25 |
|---|


















علی –
با کیفیت
پرند –
باسلام، خیلی راضی بودم ممنون از قیمت های مناسب تان